Human Milk Oligosaccharides and the Infant Gut Microbiome
University of Illinois at Urbana-Champaign Illinois Transdisciplinary Obesity Prevention Program (I-TOPP)
Urbana, IL, USA
sdonovan@illinois.edu
- Human milk oligosaccharides (HMOs) are diverse, biologically active components that benefi cially modulate the infant microbiota as well as gut, immune, and potentially neurological development. It is now possible to produce large quantities of HMOs enabling supplementation to infant formula, with the goal of supporting the gut microbiota composition and developmental outcomes more similar to that of the breastfed infant.
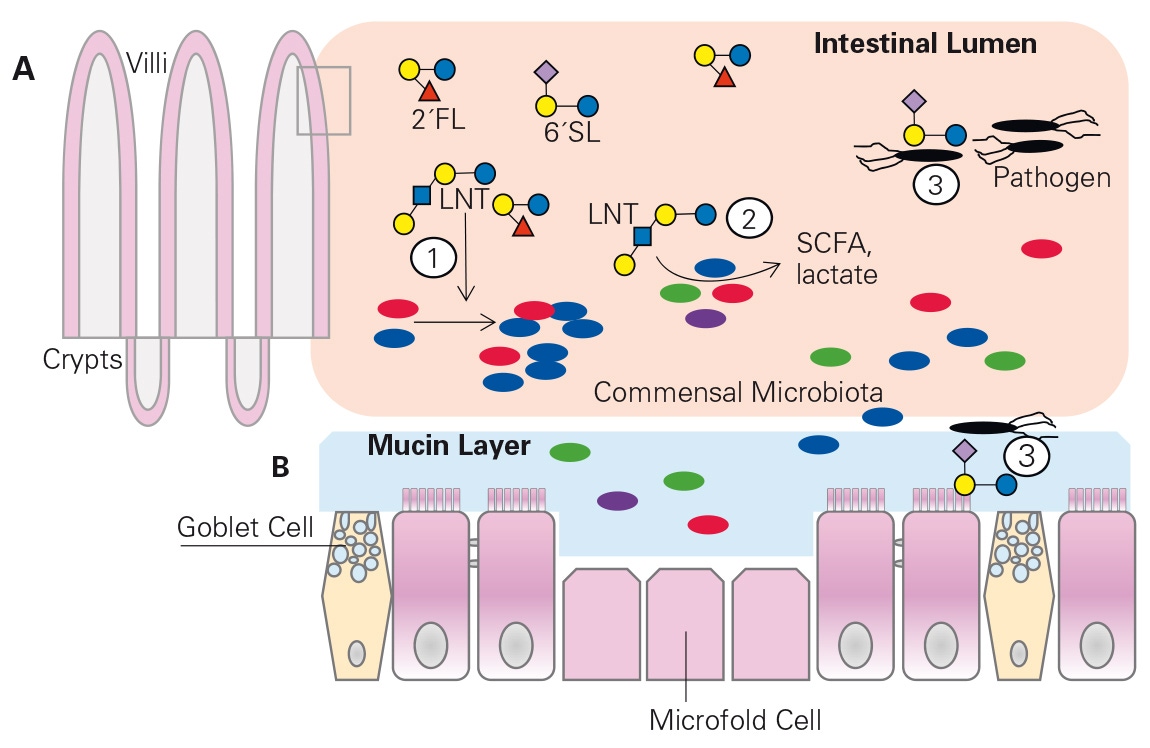
HMOs are resistant to digestion and infl uence the composition of the infant gut microbiome in several ways: by serving as prebiotics, by acting as substrates for fermentation to short-chain fatty acids, and by reducing pathogens (fi g. 1) [3, 4]. The gut microbiota of breastfed infants are typically dominated by bifi dobacterial species, with a unique enrichment of Bifi dobacterium longum spp. infantis or B. infantis [2, 5]. Most bifi dobacterial species that grow on HMOs only metabolize one of the predomi -nant HMOs, namely lacto-Ntetraose, whereas B. infantis grows well on several HMOs [6]. Genome sequencing identifi ed that B. infantis is unique in that it contains all of the oligosaccharide transport proteins and enzymes needed to transport intact HMOs into the cell, where it is broken down internally [7]. In contrast, other bifi dobacterial [8] and Bacteroides species [9] have the enzymes that break down the HMOs on their outer cell membrane and then transport the products into the cell for metabolism [7, 8]. If the HMO is hydrolyzed outside the cell, then other bacteria have access to these sugar compounds, which is referred to as cross-feeding [8]. Indeed, different HMOs in milk are both positively and negatively correlated with a number of bacteria in the stool of breastfed infants. Thus, HMOs have broad effects on shaping the infant microbiome, and it is possible to identify which HMO types are prebiotics for specifi c bacteria in the infant’s stool [2, 10].
- Li M, Wang M, Donovan SM: Early development of the gut microbiome and immune-mediated childhood disorders. Semin Reprod Med 2014;32:74–86.
- Wang M, Li M, Wu S, Lebrilla CB, Chapkin RS, Ivanov I, Donovan SM: Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J Pediatr Gastroenterol Nutr 2015;60:825–833.
- Bode L: The functional biology of human milk oligosaccharides. Early Hum Dev 2015;91:619–622.
- Jost T, Lacroix C, Braegger C, Chassard C: Impact of human milk bacteria and oligosaccharides on neonatal gut microbiota establishment and gut health. Nutr Rev 2015;73:426–437.
- Matsuki T Watanabe K, Tanaka R: Genus- and species-specifi c PCR primers for the detection and identifi cation of bifi dobacteria. Curr Issues Intest Microbiol 2003;4:61–69.
- Barboza M, Sela DA, Pirim C, Locascio RG, Freeman SL, German JB, Mills DA, Lebrilla CB: Glycoprofi ling bifi dobacterial consumption of galacto-oligosaccharides by mass spectrometry reveals strain-specifi c, preferential consumption of glycans. Appl Environ Microbiol 2009;75:7319–7325.
- Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA: The genome sequence of Bifi dobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA 2008;105:18964–18969.
- Garrido D, Barile D, Mills DA: A molecular basis for bifi dobacterial enrichment in the infant gastrointestinal tract. Adv Nutr 2012;3:415S–421S.
- Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL: Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514.
- Chichlowski M, German JB, Lebrilla CB, Mills DA: The infl uence of milk oligosaccharides on microbiota of infants: opportunities for formulas. Annu Rev Food Sci Technol 2011;2:331–351.
